FISHES aims to integrate Genomics and Fisheries Science with Indigenous Knowledge to address critical socio-economic challenges and opportunities related to food security and commercial, recreational and subsistence fisheries. This project is funded through the competitive grant from Genome Canada (Large-Scale Applied Research Project Competition: Genomic Solutions for Agriculture, Agri-Food, Fisheries and Aquaculture)
Why FISHES project?
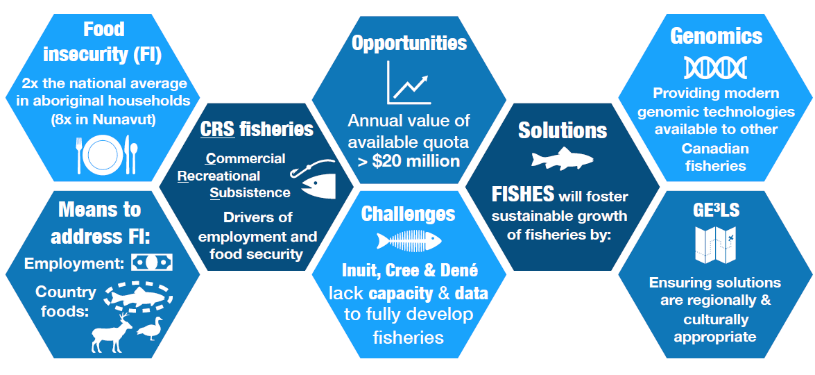
 In Canadian north, communities depend on local fisheries for both food security and cultural reasons. However, food insecurity in Northern Canada is twice as high as the national average and is even higher in Nunavut. Moreover, warming in Northern Canada is occurring at twice the rate of the rest of the planet. Increased shipping, tourism, and mining are other serious risks for northern aquatic biodiversity and fisheries.
In Canadian north, communities depend on local fisheries for both food security and cultural reasons. However, food insecurity in Northern Canada is twice as high as the national average and is even higher in Nunavut. Moreover, warming in Northern Canada is occurring at twice the rate of the rest of the planet. Increased shipping, tourism, and mining are other serious risks for northern aquatic biodiversity and fisheries.
As northern fisheries undergo these major changes, it is crucial to find ways to mitigate the negative impacts on communities of Canadian north that depend on freshwater and anadromous fish for both consumption and cultural life. Despite increased spending on food support programs, there remains an urgent social challenge surrounding these fisheries.
Threats and Challenges
Northern fisheries do not benefit from the same cutting-edge scientific information available in southern Canada. There is insufficient scientific data to facilitate sustainable commercial, recreational, and subsistence (CRS) fisheries management practices in this rapidly changing environment. As a result, improved knowledge on fish stocks, management practices, and governance is urgently needed. Identifying regions with potential for subsistence, commercial, and recreational fishing and whether they comprise genetically distinct populations is a key requirement for adaptive harvesting.
The FISHES solutions
The FISHES project works with our Indigenous partners to create an ethical, responsible, and culturally appropriate space for dialogue and shared learning for fisheries genomics research. Our research objectives are regurlarly discussed and controlled by regional research advisory boards. Using a novel knowledge co-evolution framework (for more details), we will connect Genomics and Fisheries Science with Indigenous Knowledge (IK) in support of sustainable harvests of the most preferred and beneficial local use of fisheries for subsistence, recreational and commercial harvesting. We will develop genomic resources for seven species important to communities of Canadian north (Inuit, Cree, Dené) and use these resources to identify genetically distinct populations, assess their vulnerability to future climatic conditions, quantify their contribution to mixed-population harvests, and evaluate the contribution of fish from developing hatchery programs to subsistence harvests and sustainability of fish stocks.
FISHES is built around five integrated research activities:
1- Development of genomic resources for genotyping
2- Document population structure and local adaptation
3- Genomics bio-monitoring for maintaining fish food security in strategic northern regions
4- Assessing genomic signals of selection to predict fish stock vulnerability to climate change
5- Combining genomics, bio-monitoring and IK for inclusive fisheries co-management, and to enhance food security and socio-economic development in northern Indigenous community settings
Deliverables
- Reduce knowledge inequities and power imbalances by providing cutting-edge scientific information to manage CRS fisheries throughout Northern Canada.
- Enhance food security for Canada’s northern Indigenous people by providing access to nutritious and culturally-preferred foods.
- Generate economic value and jobs through new aquaculture development opportunities, commercial fishery activities, and processing plants.
- Forecast the response of key fisheries resources to rapid global changes and socio-economic changes in Canada’s northern Indigenous communities.
- Create unique opportunities for comparative genomics, fisheries science and IK research synergies.

















